Poisson regression example
Contents
8. Poisson regression example#
8.1. Modeling tropical storm counts#
The number of tropical storms is an example of a variable that is not expected to be normally distributed. Counts cannot be negative, are often skewed and tend to follow a Poisson distribution.
This example follows the general approach of Villarini et al. (2010) in using Poisson regression to model the number of tropical storms based on climate indices.
The goal of Poisson regression is to model a rate of occurrence, \(\Lambda_i\), where the rate changes for each observation \(i\). This model can be expressed as a generalized linear model (GLM) with a logarithmic link function.
which is the same as
This is a model for the rate of occurrence \(\Lambda_i\) as a function of \(k\) predictor variables. In this example, the rate of occurrence \(\Lambda_i\) is the number of storm counts per year and each climate index is a predictor variable.
The logarithmic link function is useful because \(\log(\Lambda_i)\) can be positive or negative, but \(\Lambda_i\) must be positive
If the data being modeled is a standard Poisson random variable, the model simplifies to \(\Lambda_i = \exp({\beta_0})\)
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from scipy import stats
import statsmodels.api as sm
import statsmodels.formula.api as smf
We use a special function to load the climate indices from NOAA.
def read_psl_file(psl_file):
'''
Read a data file in in NOAA Physical Sciences Laboratory (PSL) format
Input: String containing the path to a PSL data file
Returns: Pandas dataframe with monthly data in columns and the year as the index
For a description of the PSL format see: https://psl.noaa.gov/gcos_wgsp/Timeseries/standard/
'''
f = open(psl_file, "r")
all_lines = f.readlines()
start_year = all_lines[0].split()[0]
end_year = all_lines[0].split()[1]
for i in range(1,len(all_lines)):
stri = all_lines[i].find(end_year)
if stri>=0:
end_index = i
missing_val = float(all_lines[end_index+1])
nrows = int(end_year)-int(start_year)+1
df = pd.read_csv(psl_file,skiprows=1,nrows=nrows,sep='\s+',header=None,na_values=missing_val)
df = df.rename(columns={0:'year'})
df = df.set_index('year',drop=True)
return df
dfsoi = read_psl_file('data/tropical-storms/soi.data')
dftna = read_psl_file('data/tropical-storms/tna.data')
dfnao = read_psl_file('data/tropical-storms/nao.data')
Load the tropical storm data.
dftrop = pd.read_csv('data/tropical-storms/tropical.txt',sep='\t')
dftrop = dftrop.set_index('Year',drop=False)
Tropical storms do not happen in winter, so we average the climate indices during the relevant months.
dftrop['SOI'] = dfsoi.loc[:,5:6].mean(axis=1)
dftrop['TNA'] = dftna.loc[:,5:6].mean(axis=1)
dftrop['NAO'] = dfnao.loc[:,5:6].mean(axis=1)
Exercises#
How would you find the NAO averaged over July-September?
How would you find the NAO for years 2010-2020, averaged over July-September?
How would you use
dfnao.ilocto achieve the same result?
Drop all rows with any NaN values.
df = dftrop.dropna()
df
| Year | NamedStorms | NamedStormDays | Hurricanes | HurricaneDays | MajorHurricanes | MajorHurricaneDays | AccumulatedCycloneEnergy | SOI | TNA | NAO | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | |||||||||||
| 1951 | 1951 | 12 | 67.00 | 8 | 34.25 | 3 | 4.50 | 126.3 | -0.40 | 0.155 | -0.925 |
| 1952 | 1952 | 11 | 45.75 | 5 | 15.50 | 2 | 2.50 | 69.1 | 1.20 | 0.205 | -0.545 |
| 1953 | 1953 | 14 | 61.75 | 7 | 19.00 | 3 | 5.00 | 98.5 | -1.30 | 0.120 | 0.425 |
| 1954 | 1954 | 16 | 57.00 | 7 | 26.00 | 3 | 7.00 | 104.4 | 0.50 | 0.045 | 0.015 |
| 1955 | 1955 | 13 | 85.50 | 9 | 43.00 | 4 | 8.50 | 164.7 | 1.95 | -0.065 | -0.530 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2016 | 2016 | 15 | 81.00 | 7 | 27.75 | 4 | 10.25 | 141.3 | 0.90 | 0.385 | -0.400 |
| 2017 | 2017 | 17 | 93.00 | 10 | 51.75 | 6 | 19.25 | 224.9 | -0.15 | 0.590 | -0.685 |
| 2018 | 2018 | 15 | 87.25 | 8 | 26.75 | 2 | 5.00 | 129.0 | 0.20 | -0.440 | 1.715 |
| 2019 | 2019 | 18 | 68.50 | 6 | 23.25 | 3 | 10.00 | 129.8 | -0.70 | 0.210 | -1.585 |
| 2020 | 2020 | 30 | 118.00 | 13 | 34.75 | 6 | 8.75 | 179.8 | 0.05 | 0.620 | -0.085 |
70 rows × 11 columns
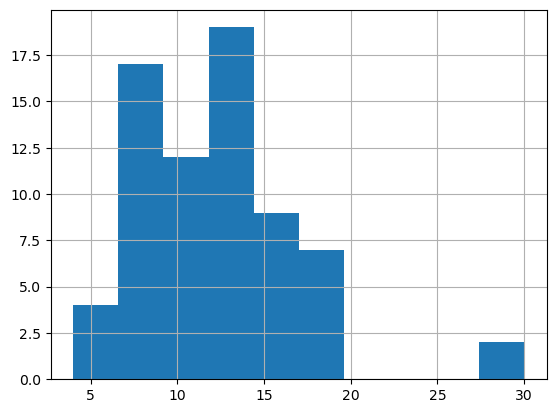
Histogram of the named storm counts.
plt.figure()
df['NamedStorms'].hist()
<AxesSubplot:>
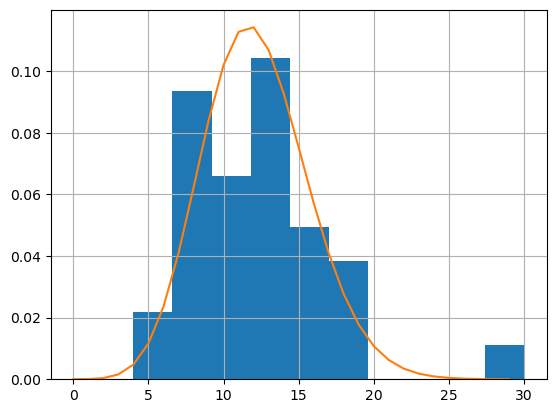
Poisson distribution - model as a constant
result = smf.glm(formula='NamedStorms ~ 1',
data=df,
family=sm.families.Poisson()).fit()
result.summary()
| Dep. Variable: | NamedStorms | No. Observations: | 70 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 69 |
| Model Family: | Poisson | Df Model: | 0 |
| Link Function: | Log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -206.92 |
| Date: | Mon, 13 Jan 2025 | Deviance: | 113.91 |
| Time: | 11:28:25 | Pearson chi2: | 123. |
| No. Iterations: | 4 | Pseudo R-squ. (CS): | -1.554e-15 |
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 2.4979 | 0.034 | 72.869 | 0.000 | 2.431 | 2.565 |
k = np.arange(0,30)
mu = np.exp(result.params.Intercept)
pmf_poisson = stats.poisson.pmf(k,mu)
plt.figure()
df['NamedStorms'].hist(density=True)
plt.plot(k,pmf_poisson)
[<matplotlib.lines.Line2D at 0x308c6b700>]
Poisson regression model, including a linear temporal trend.
result = smf.glm(formula='NamedStorms ~ Year + NAO + SOI + TNA',
data=df,
family=sm.families.Poisson()).fit()
result.summary()
| Dep. Variable: | NamedStorms | No. Observations: | 70 |
|---|---|---|---|
| Model: | GLM | Df Residuals: | 65 |
| Model Family: | Poisson | Df Model: | 4 |
| Link Function: | Log | Scale: | 1.0000 |
| Method: | IRLS | Log-Likelihood: | -186.77 |
| Date: | Mon, 13 Jan 2025 | Deviance: | 73.614 |
| Time: | 11:28:25 | Pearson chi2: | 73.3 |
| No. Iterations: | 4 | Pseudo R-squ. (CS): | 0.4377 |
| Covariance Type: | nonrobust |
| coef | std err | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -8.9786 | 3.621 | -2.480 | 0.013 | -16.076 | -1.881 |
| Year | 0.0057 | 0.002 | 3.154 | 0.002 | 0.002 | 0.009 |
| NAO | -0.0267 | 0.048 | -0.558 | 0.577 | -0.120 | 0.067 |
| SOI | 0.0620 | 0.037 | 1.654 | 0.098 | -0.011 | 0.135 |
| TNA | 0.3563 | 0.096 | 3.729 | 0.000 | 0.169 | 0.544 |
beta = result.params
beta
Intercept -8.978558
Year 0.005747
NAO -0.026677
SOI 0.061967
TNA 0.356328
dtype: float64
Exercises#
Create a Python variable
ythat contains the observed count data.Create a Python variable
yhatthat contains the modeled count data (use the parameters inbeta).Make plots comparing the observed and modeled counts.