Cruise data analysis in Python
Contents
4. Cruise data analysis in Python#
4.1. WCOA 2013 data set#
To download the data used in this tutorial, use the following command in the Terminal (Mac) or Git Bash (Windows).
git clone https://github.com/mlmldata24/wcoa_cruise.git
The data comes from the West Coast Ocean Acidification (WCOA) cruise in 2013. The goal of this NOAA-supported research cruise is to collect data to help understand the effects of coastal upwelling on ocean acidification, and the impacts of ocean acidification on organisms and ecosystems. This video gives an idea of life aboard the ship and the type of science operations conducted.
In this part of the tutorial, we will go over the basics of working with dates in Pandas and Numpy, make some exploratory plots and start a regression analysis. The data exploration will be largely guided by student interest.
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from scipy import stats
Introduction to Pandas dataframes#
We use Pandas to import the csv data file.
Here, there is an optional parse_dates argument. The numbers in double brackets [[8,9]] indicate which columns to interpret as dates.
filename = 'data/wcoa_cruise/WCOA2013_hy1.csv'
df = pd.read_csv(filename,header=31,na_values=-999,
parse_dates=[[8,9]])
df.head()
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2013-08-05 02:12:20 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 1 | 2 | 48.2 | ... | 2370.2 | 2 | 2369.0 | 2 | 7.294 | 2 | 25.0 | NaN | 9 | NaN |
| 1 | 2013-08-05 02:12:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 2 | 2 | 48.2 | ... | NaN | 9 | NaN | 9 | 7.295 | 2 | 25.0 | NaN | 9 | NaN |
| 2 | 2013-08-05 02:19:58 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 3 | 2 | 48.2 | ... | 2349.6 | 2 | 2343.7 | 2 | 7.282 | 2 | 25.0 | 43.521 | 3 | NaN |
| 3 | 2013-08-05 02:27:01 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 4 | 2 | 48.2 | ... | 2318.7 | 2 | 2311.9 | 2 | 7.287 | 2 | 25.0 | 45.641 | 2 | NaN |
| 4 | 2013-08-05 02:30:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 5 | 2 | 48.2 | ... | 2300.0 | 2 | 2299.7 | 2 | 7.308 | 2 | 25.0 | 47.741 | 2 | NaN |
5 rows × 42 columns
df.columns
Index(['DATE_TIME', 'EXPOCODE', 'SECT_ID', 'LEG', 'LINE', 'STNNBR', 'CASTNO',
'BTLNBR', 'BTLNBR_FLAG_W', 'LATITUDE', 'LONGITUDE', 'DEPTH', 'CTDPRS',
'CTDTMP', 'CTDSAL', 'CTDSAL_FLAG_W', 'CTDOXY', 'CTDOXY_FLAG_W',
'SALNTY', 'SALNTY_FLAG_W', 'OXYGEN', 'OXYGEN_FLAG_W', 'SILCAT',
'SILCAT_FLAG_W', 'NITRAT', 'NITRAT_FLAG_W', 'NITRIT', 'NITRIT_FLAG_W',
'PHSPHT', 'PHSPHT_FLAG_W', 'AMMONI', 'AMMONI_FLAG_W', 'TCARBN',
'TCARBN_FLAG_W', 'ALKALI', 'ALKALI_FLAG_W', 'PH_TOT', 'PH_TOT_FLAG_W',
'PH_TMP', 'CO32', 'CO32__FLAG_W', 'CHLORA'],
dtype='object')
Instead of strings, the dates are now in a special datetime64 format. This means that, instead of treating the dates in the same way as any other collection of characters, pandas and NumPy can understand how this variable represents time.
df['DATE_TIME'].head()
0 2013-08-05 02:12:20
1 2013-08-05 02:12:53
2 2013-08-05 02:19:58
3 2013-08-05 02:27:01
4 2013-08-05 02:30:53
Name: DATE_TIME, dtype: datetime64[ns]
For example, subtracting datetime64 objects with pandas gives a Timedelta object, which is specifically used to represent differences between times. The first two samples in the cruise data are separated by 33 seconds (the time between firing of bottles).
df['DATE_TIME'][1]-df['DATE_TIME'][0]
Timedelta('0 days 00:00:33')
pd.unique(df['LATITUDE'])
array([48.2 , 48.3 , 48.37, 48.44, 48.5 , 48.53, 48.61, 48.66, 48.71,
48.78, 48.81, 48.84, 47.97, 48.14, 47.96, 47.68, 47.13, 47.11,
47.12, 47.34, 46.13, 46.17, 46.19, 46.25, 46.24, 46.12, 44.65,
44.66, 44.2 , 41.99, 41.97, 41.96, 41.94, 41.9 , 40.25, 40.23,
40.22, 40.21, 40.1 , 37.67, 37.94, 37.91, 37.87, 37.76, 37.75,
36.8 , 36.78, 36.76, 36.73, 36.71, 36.69, 36.52, 36.7 ])
Exercise#
Create a list of unique station ID’s (“STNNBR”) found in the survey data. Call it stns. How many unique stations are there in the data?
Summary statistics#
A summary of the dataframe is given by the .describe() method.
df['CTDTMP'].describe()
count 969.000000
mean 8.993954
std 3.055917
min 1.738200
25% 7.292000
50% 8.328200
75% 10.752200
max 20.747400
Name: CTDTMP, dtype: float64
These summary statistics can also be accessed individually with similar syntax.
df['CTDTMP'].mean()
8.993954179566563
df['CTDTMP'].min()
1.7382
Alternate method using Numpy functions.
np.min(df['CTDTMP'])
1.7382
Mathematical operations#
Converting Celcius to Fahrenheit
df['CTDTMP_F'] = 9/5*df['CTDTMP'] + 32
df['CTDTMP_F'].head()
0 38.63894
1 38.64236
2 39.85916
3 41.05328
4 41.82404
Name: CTDTMP_F, dtype: float64
df['CTDTMP'].head()
0 3.6883
1 3.6902
2 4.3662
3 5.0296
4 5.4578
Name: CTDTMP, dtype: float64
Plotting#
Plot latitude as a function of time.
plt.figure()
plt.plot(df['DATE_TIME'],df['LATITUDE'],'-o')
plt.xticks(rotation=15)
(array([15922., 15926., 15930., 15934., 15938., 15942., 15946.]),
[Text(0, 0, ''),
Text(0, 0, ''),
Text(0, 0, ''),
Text(0, 0, ''),
Text(0, 0, ''),
Text(0, 0, ''),
Text(0, 0, '')])
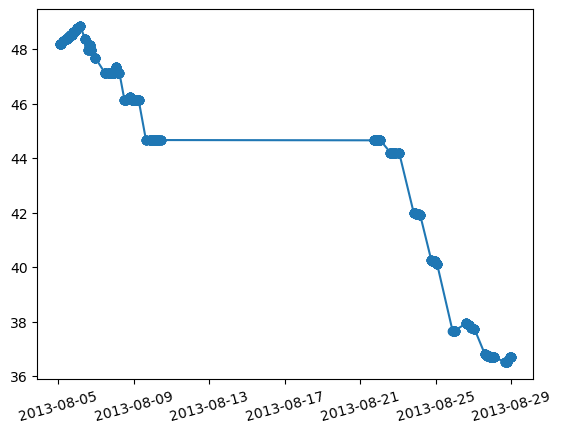
The pyplot library automatically understands datetime64 objects so it is easy to see how the ship moved between stations from north to south as weeks passed.
plt.figure()
plt.plot(df['LONGITUDE'], df['LATITUDE'], 'ro')
[<matplotlib.lines.Line2D at 0x1697cf850>]
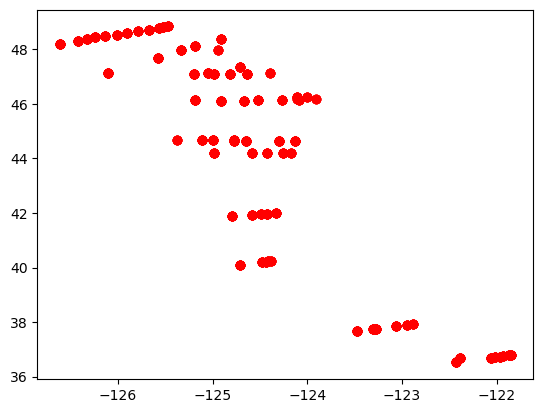
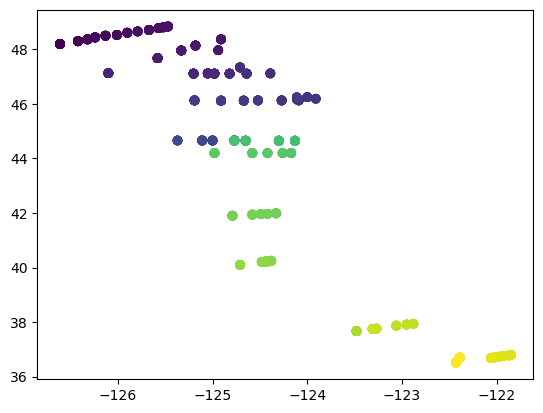
The scatter() function allows points to be colored according to the value of a variable. In the case of dates, later dates are shown as warmer colors.
plt.figure()
plt.scatter(df['LONGITUDE'],df['LATITUDE'],c=df['DATE_TIME'])
<matplotlib.collections.PathCollection at 0x169846da0>
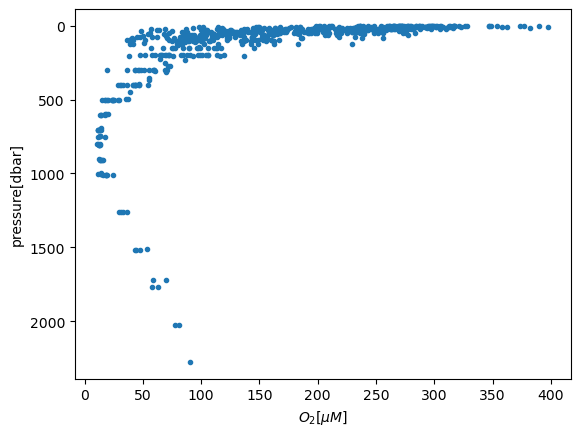
Note that the vertical coordinate is pressure (not depth, which indicates the bottom depth rather than the depth of the sample). To plot dissolved oxygen with depth:
plt.figure()
plt.plot(df['OXYGEN'],df['CTDPRS'],'.')
plt.gca().invert_yaxis()
plt.xlabel('$O_2 [\mu M]$')
plt.ylabel('pressure[dbar]')
Text(0, 0.5, 'pressure[dbar]')
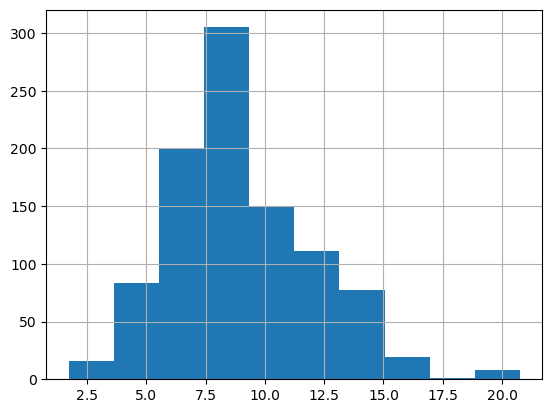
plt.figure()
df['CTDTMP'].hist()
<AxesSubplot:>
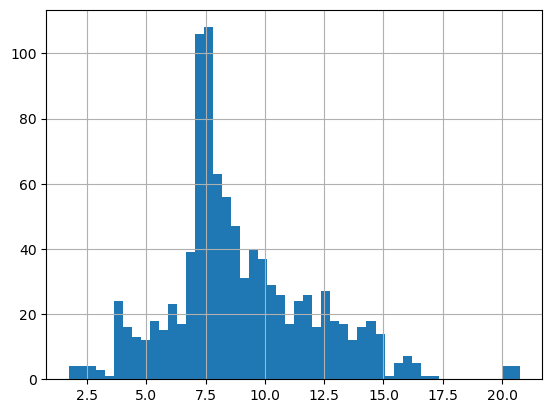
plt.figure()
df['CTDTMP'].hist(bins=50)
<AxesSubplot:>
df.keys()
Index(['DATE_TIME', 'EXPOCODE', 'SECT_ID', 'LEG', 'LINE', 'STNNBR', 'CASTNO',
'BTLNBR', 'BTLNBR_FLAG_W', 'LATITUDE', 'LONGITUDE', 'DEPTH', 'CTDPRS',
'CTDTMP', 'CTDSAL', 'CTDSAL_FLAG_W', 'CTDOXY', 'CTDOXY_FLAG_W',
'SALNTY', 'SALNTY_FLAG_W', 'OXYGEN', 'OXYGEN_FLAG_W', 'SILCAT',
'SILCAT_FLAG_W', 'NITRAT', 'NITRAT_FLAG_W', 'NITRIT', 'NITRIT_FLAG_W',
'PHSPHT', 'PHSPHT_FLAG_W', 'AMMONI', 'AMMONI_FLAG_W', 'TCARBN',
'TCARBN_FLAG_W', 'ALKALI', 'ALKALI_FLAG_W', 'PH_TOT', 'PH_TOT_FLAG_W',
'PH_TMP', 'CO32', 'CO32__FLAG_W', 'CHLORA', 'CTDTMP_F'],
dtype='object')
Exercises#
What scientific questions can be addressed with this data set?
What relationships might occur between different variables?
What differences might occur within the same variables, but at different locations or times?
Create exploratory plots (one PDF, one scatter plot)
4.2. Slicing and subsetting data#
df[0:3]
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2013-08-05 02:12:20 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 1 | 2 | 48.2 | ... | 2 | 2369.0 | 2 | 7.294 | 2 | 25.0 | NaN | 9 | NaN | 38.63894 |
| 1 | 2013-08-05 02:12:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 2 | 2 | 48.2 | ... | 9 | NaN | 9 | 7.295 | 2 | 25.0 | NaN | 9 | NaN | 38.64236 |
| 2 | 2013-08-05 02:19:58 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 3 | 2 | 48.2 | ... | 2 | 2343.7 | 2 | 7.282 | 2 | 25.0 | 43.521 | 3 | NaN | 39.85916 |
3 rows × 43 columns
df.iloc[0:3]
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2013-08-05 02:12:20 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 1 | 2 | 48.2 | ... | 2 | 2369.0 | 2 | 7.294 | 2 | 25.0 | NaN | 9 | NaN | 38.63894 |
| 1 | 2013-08-05 02:12:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 2 | 2 | 48.2 | ... | 9 | NaN | 9 | 7.295 | 2 | 25.0 | NaN | 9 | NaN | 38.64236 |
| 2 | 2013-08-05 02:19:58 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 3 | 2 | 48.2 | ... | 2 | 2343.7 | 2 | 7.282 | 2 | 25.0 | 43.521 | 3 | NaN | 39.85916 |
3 rows × 43 columns
df.loc[0:3]
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2013-08-05 02:12:20 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 1 | 2 | 48.2 | ... | 2 | 2369.0 | 2 | 7.294 | 2 | 25.0 | NaN | 9 | NaN | 38.63894 |
| 1 | 2013-08-05 02:12:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 2 | 2 | 48.2 | ... | 9 | NaN | 9 | 7.295 | 2 | 25.0 | NaN | 9 | NaN | 38.64236 |
| 2 | 2013-08-05 02:19:58 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 3 | 2 | 48.2 | ... | 2 | 2343.7 | 2 | 7.282 | 2 | 25.0 | 43.521 | 3 | NaN | 39.85916 |
| 3 | 2013-08-05 02:27:01 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 4 | 2 | 48.2 | ... | 2 | 2311.9 | 2 | 7.287 | 2 | 25.0 | 45.641 | 2 | NaN | 41.05328 |
4 rows × 43 columns
df.loc[0:3,['CTDTMP','CTDPRS']]
| CTDTMP | CTDPRS | |
|---|---|---|
| 0 | 3.6883 | 999.5 |
| 1 | 3.6902 | 1000.8 |
| 2 | 4.3662 | 749.0 |
| 3 | 5.0296 | 503.9 |
Exercise#
What do you expect to happen when you execute:
df[0:1]
df[:4]
df[:-1]
What do you expect to happen when you call:
df.iloc[0:4, 1:4]
df.loc[0:4, 1:4]
How are the two commands different?
Adapted from: https://datacarpentry.org/python-ecology-lesson/03-index-slice-subset/index.html
Subsetting Data using Criteria#
df[df.LATITUDE > 40].head()
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2013-08-05 02:12:20 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 1 | 2 | 48.2 | ... | 2 | 2369.0 | 2 | 7.294 | 2 | 25.0 | NaN | 9 | NaN | 38.63894 |
| 1 | 2013-08-05 02:12:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 2 | 2 | 48.2 | ... | 9 | NaN | 9 | 7.295 | 2 | 25.0 | NaN | 9 | NaN | 38.64236 |
| 2 | 2013-08-05 02:19:58 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 3 | 2 | 48.2 | ... | 2 | 2343.7 | 2 | 7.282 | 2 | 25.0 | 43.521 | 3 | NaN | 39.85916 |
| 3 | 2013-08-05 02:27:01 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 4 | 2 | 48.2 | ... | 2 | 2311.9 | 2 | 7.287 | 2 | 25.0 | 45.641 | 2 | NaN | 41.05328 |
| 4 | 2013-08-05 02:30:53 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 5 | 2 | 48.2 | ... | 2 | 2299.7 | 2 | 7.308 | 2 | 25.0 | 47.741 | 2 | NaN | 41.82404 |
5 rows × 43 columns
df[df.LATITUDE <= 40].head()
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 770 | 2013-08-25 21:23:35 | 32P020130821 | WCOA2013 | 2 | 10 | 133 | 1 | 1 | 2 | 37.67 | ... | 6 | 2430.4 | 6 | 7.493 | 2 | 25.0 | 73.346 | 2 | NaN | 35.12876 |
| 771 | 2013-08-25 21:32:13 | 32P020130821 | WCOA2013 | 2 | 10 | 133 | 1 | 2 | 2 | 37.67 | ... | 2 | 2427.6 | 2 | 7.467 | 2 | 25.0 | 69.812 | 2 | NaN | 35.30336 |
| 772 | 2013-08-25 21:40:44 | 32P020130821 | WCOA2013 | 2 | 10 | 133 | 1 | 3 | 2 | 37.67 | ... | 2 | 2424.3 | 2 | 7.442 | 2 | 25.0 | 65.502 | 2 | NaN | 35.55914 |
| 773 | 2013-08-25 21:49:45 | 32P020130821 | WCOA2013 | 2 | 10 | 133 | 1 | 4 | 2 | 37.67 | ... | 2 | 2413.7 | 2 | 7.400 | 2 | 25.0 | 60.170 | 2 | NaN | 36.10526 |
| 774 | 2013-08-25 21:57:55 | 32P020130821 | WCOA2013 | 2 | 10 | 133 | 1 | 5 | 2 | 37.67 | ... | 2 | 2401.4 | 2 | 7.374 | 2 | 25.0 | 57.739 | 2 | NaN | 36.92894 |
5 rows × 43 columns
dfsub = df[(df.CTDPRS <= 10) & (df.LATITUDE > 40)]
dfsub.head()
| DATE_TIME | EXPOCODE | SECT_ID | LEG | LINE | STNNBR | CASTNO | BTLNBR | BTLNBR_FLAG_W | LATITUDE | ... | TCARBN_FLAG_W | ALKALI | ALKALI_FLAG_W | PH_TOT | PH_TOT_FLAG_W | PH_TMP | CO32 | CO32__FLAG_W | CHLORA | CTDTMP_F | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 2013-08-05 03:00:52 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 19 | 4 | 48.20 | ... | 9 | NaN | 9 | NaN | 9 | NaN | NaN | 9 | NaN | 56.54660 |
| 19 | 2013-08-05 03:01:10 | 317W20130803 | WCOA2013 | 1 | 2 | 11 | 1 | 20 | 2 | 48.20 | ... | 2 | 2189.1 | 6 | 7.983 | 3 | 25.0 | 155.043 | 2 | NaN | 56.54570 |
| 38 | 2013-08-05 06:37:22 | 317W20130803 | WCOA2013 | 1 | 2 | 12 | 1 | 19 | 2 | 48.30 | ... | 9 | 2180.0 | 6 | 7.980 | 3 | 25.0 | 152.868 | 2 | NaN | 57.72218 |
| 39 | 2013-08-05 06:37:42 | 317W20130803 | WCOA2013 | 1 | 2 | 12 | 1 | 20 | 2 | 48.30 | ... | 2 | NaN | 9 | 7.981 | 2 | 25.0 | NaN | 5 | NaN | 57.72290 |
| 58 | 2013-08-05 10:41:19 | 317W20130803 | WCOA2013 | 1 | 2 | 13 | 1 | 19 | 2 | 48.37 | ... | 2 | 2178.7 | 2 | 7.931 | 2 | 25.0 | 142.629 | 2 | NaN | 55.69844 |
5 rows × 43 columns
Exercises#
Select a subset of rows in the
dfDataFrame that contains data from a pressure range between 500 and 1000 dbar. How many rows did you end up with? What did your neighbor get?You can use the isin command in Python to query a DataFrame based upon a list of values as follows:
df[df['STNNBR'].isin([listGoesHere])]
Use the isin function to find all samples from station numbers 11 and 12.
Exercises#
pH values are contained in the df['PH_TOT'] DataArray
Quality control flags for pH are contained in the df['PH_TOT_FLAG_W'] DataArray.
World Ocean Circulation Experiment (WOCE) quality control flags are used:
2 = good value
3 = questionable value
4 = bad value
5 = value not reported
6 = mean of replicate measurements
9 = sample not drawn.
Create a new DataFrame called
dfsub1that includes only data where pressure is less than 10 dbar and the latitude is farher north than \(40^{\circ} \text{N}\).
dfsub = df[(df['CTDPRS'] > 10) & (df['LATITUDE'] > 40)]
Create a new DataFrame called
dfsub2that excludes all bad, questionable and missing pH and CTD oxygen values. Plot oxygen vs. pH.
dfsub2 = df[(df['CTDOXY_FLAG_W'] == 2) & (df['PH_TOT_FLAG_W'] == 2)]
Fit a linear model in Python#
plt.plot(df['CTDOXY'],df['PH_TOT'],'.')
plt.xlabel('dissolved oxygen [$\mu$mol/kg]')
plt.ylabel('pH$_{Tot}$')
Text(0, 0.5, 'pH$_{Tot}$')
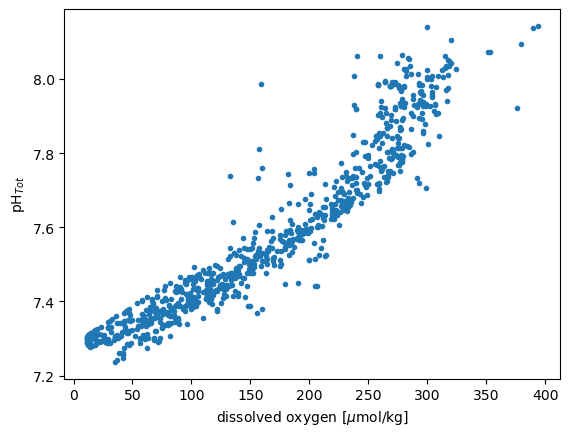
result = stats.linregress(dfsub2['CTDOXY'],dfsub2['PH_TOT'])
result
LinregressResult(slope=0.002305076546103237, intercept=7.202036145940105, rvalue=0.9510631019969654, pvalue=0.0, stderr=2.7737424376777354e-05, intercept_stderr=0.004977802641597415)
Exercise#
Plot the linear model with the data.